| Citation: | CHEN Xiao-Hui, ZHOU Guo-Liang, SUN Chun-Xiao, ZHANG Xiao-min, ZHANG Guo-Jian, ZHU Tian-Jiao, LI Jing, CHE Qian, LI De-Hai. Penicacids E−G, three new mycophenolic acid derivatives from the marine-derived fungus Penicillium parvum HDN17-478 [J]. Chin J Nat Med, 2020, 18(11): 850-854. DOI: 10.1016/S1875-5364(20)60027-9 |
Mycophenolic acid, a phenyl-terpenoid derivative [1], was mainly seen in the metabolites of fungal strains of the genus Penicillium, including P. brevicompactum, P. glaucum, P. scarbum, and P. grisebrunneum etc. [2]. Since the first reported of mycophenolic acid as a secondary fungal metabolite in 1893 [3–4], the mycophenolic acid and its derivatives have attracted widespread attention because of their broad bioactivities, such as immunosuppressive, antibacterial, antifungal, antiviral, and antitumor properties [5–6].
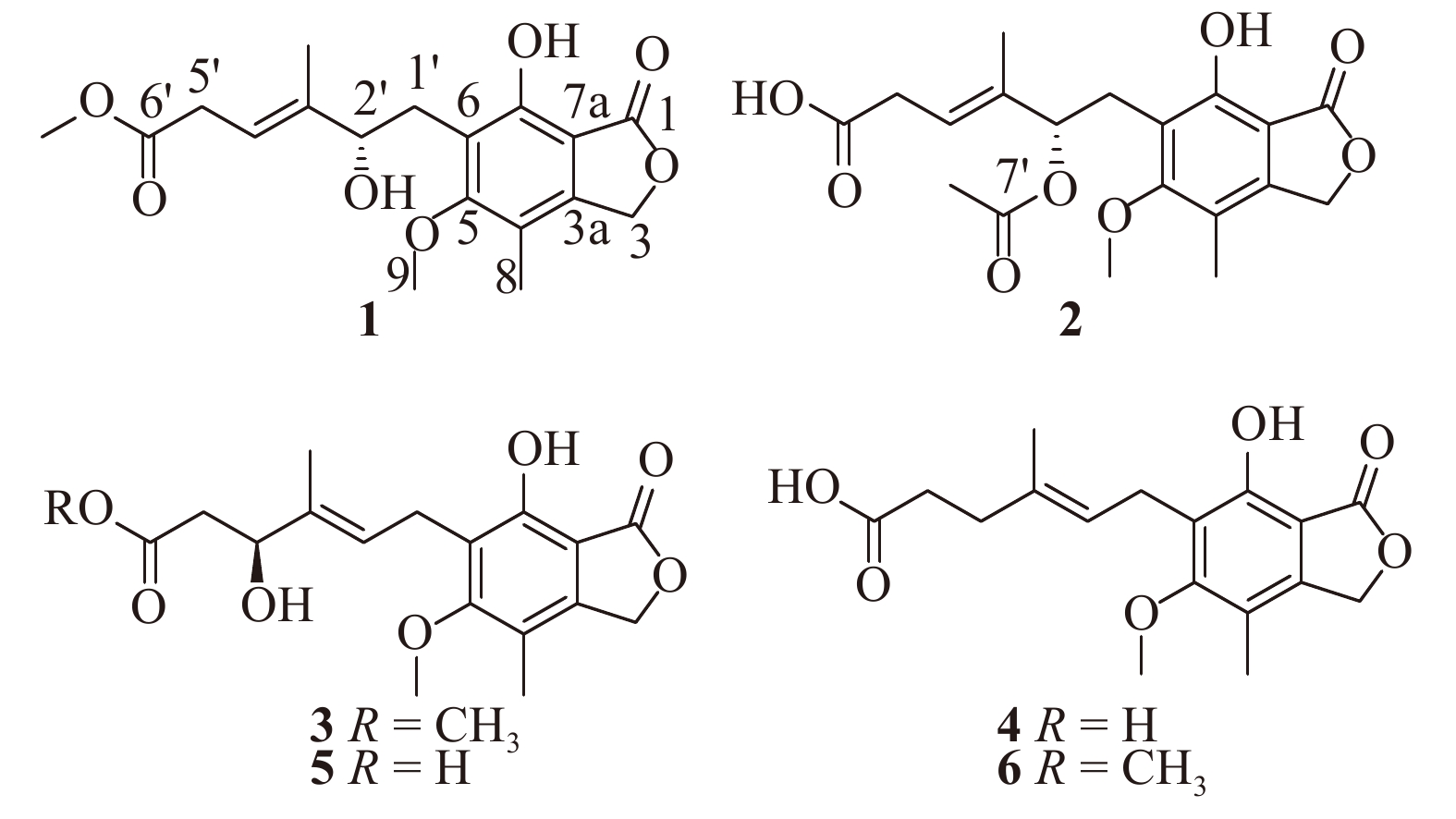
Deep-sea-derived microorganisms, with great advantage of unique ecological environments, are new potential resources for discovery of bioactive secondary metabolites [7–9]. Our previous research on deep-sea derived microorganisms has provided a series of bioactive secondary metabolites with diverse bioactivities [10–14]. For a recent work within the same scope, a fungal strain Penicillium parvum HDN17-478, isolated from a deep-sea sediment sample collected in South China Sea, was selected for its intriguing HPLC profile with characteristic UV absorption of mycophenolic acid and promising cytotoxicity of the crude extract from its ferments. Chemical investigations on the large scaled fermentation led to the isolation of three new compounds, named penicacids E−G (1−3), along with three known analogues including mycophenolic acid (4) [15], 4′-hydroxy-mycophenolic acid (5) [16], and mycophenolic methyl ester (6) [17]. To the best of our knowledge, compounds 1 and 2 possess a double bond positioned at C-3′/C-4′, which has never been seen in the members of known mycophenolic acid like natural products so far. Herein, we reported the details of the isolation, structure elucidation, absolute configuration and biological activities of the isolated compounds.
The fungal strain Penicillium parvum HDN17-478 was cultured, extracted, and subjected to repeated silica gel and semi-preparative HPLC, yielding the compounds 1 (6.4 mg), 2 (10.4 mg), 3 (8.3 mg), 4 (12.0 mg), 5 (3.2 mg), and 6 (19.3 mg) (Fig. 1).
Penicacid E (1) was obtained as a white amorphous solid, and the molecular formula was determined to be C18H22O7 by the HR-ESI-MS (m/z 373.1253 [M + Na]+), suggesting eight degrees of unsaturation. The 1H NMR data of 1 revealed a allylic methyl singlet at δH 1.77 (3H, s, 3′-CH3), an aromatic methyl singlet at δH 2.17 (3H, s, C-8), and two methoxyls at δH 3.82 (3H, s, C-9) and δH 3.68 (3H, s, 6′-OCH3). The 13C NMR data of 1 revealed the presence of four sp3 methyls, three sp3 methylenes with one oxygenated (δC 70.2/δH 5.21, C-3), one sp3 methines (δC 77.1/δH 4.34, C-2′), one sp2 methines (δC 117.2/δH 5.64, C-4′), seven sp2 quaternary carbons and two carbonyls (δC 172.7, 172.8) (Table 1). These NMR data of 1 was closely similar to the known compound mycophenolic acid (4) [15] except for the slight variations in the several side chain proton and carbon signals, indicating the same skeleton of both compound structures (Fig. 1). Further, the HMBC correlation from 6′-OCH3 to C-6′ (δC 172.8) enabled us to attach the methoxy group (δH 3.68, δC 52.1) to C-6′ (Fig. 2). Moreover, the position of double bond at C-3′ (δC 140.8) /C-4′ (δC 117.2) could be determined by the COSY correlations from H-4′ (δH 5.64) to H-5′ (δH 3.09) together with the HMBC correlations from 3′-CH3 to C-3′ and C-4′, from H-4′ to C-2′, and from H-5′ to C-4′, as shown in Fig. 2. The location of a hydroxyl group at C-2′ was indicated by the chemical shift of the C-2′ methine group (δH 4.34, δC 77.1) together with the molecular composition. Thus, the planar structure of 1 was established (Fig. 2).
| No. | 1a | 2a | 3b | |||||
| δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | |||
| 1 | 172.7, C | 173.2, C | 170.1, C | |||||
| 3 | 70.2, CH2 | 5.21, s | 70.3, CH2 | 5.20, s | 68.6, CH2 | 5.24, s | ||
| 3a | 145.3, C | 144.6, C | 145.8, C | |||||
| 4 | 117.0, C | 117.0, C | 115.9, C | |||||
| 5 | 164.1, C | 163.9, C | 162.6, C | |||||
| 6 | 119.8, C | 121.4, C | 122.1, C | |||||
| 7 | 154.2, C | 153.8, C | 152.7, C | |||||
| 7a | 107.1, C | 106.7, C | 106.9, C | |||||
| 8 | 11.9, CH3 | 2.17, s | 11.8, CH3 | 2.15, s | 11.0, CH3 | 2.08, s | ||
| 9 | 61.2, CH3 | 3.82, s | 61.2, CH3 | 3.75, s | 60.6, CH3 | 3.69, s | ||
| 1′ | 30.3, CH2 | 2.92/2.99, m | 22.5, CH2 | 3.42, t (6.9) | 22.0, CH2 | 3.30, d (6.9) | ||
| 2′ | 77.1, CH | 4.34, d (7.7) | 75.0, CH | 5.53, t (7.7) | 122.8, CH | 5.36, d (6.6) | ||
| 3′ | 140.8, C | 132.6, C | 136.8, C | |||||
| 3′-CH3 | 12.6, CH3 | 1.77, s | 12.3, CH3 | 1.82, s | 11.8, CH3 | 1.70, s | ||
| 4′ | 117.2, CH | 5.64, t (6.8) | 127.1, CH | 5.60, t (6.7) | 72.5, CH | 4.20, m | ||
| 5′ | 33.4, CH2 | 3.09, d (4.9) | 38.3, CH2 | 2.63/2.75, m | 41.0, CH2 | 2.32/2.41, m | ||
| 6′ | 172.8, C | 175.0, C | 171.3, C | |||||
| 6′-OCH3 | 52.1, CH3 | 3.68, s | 51.1, CH3 | 3.51, s | ||||
| 7′ | 170.3, C | |||||||
| 7′-CH3 | 21.3, CH3 | 2.02, s | ||||||
| a Recorded in CDCl3; b Recorded in DMSO-d6 | ||||||||
The absolute stereochemistry of 1 was determined by the Mosher’s method [18]. Compound 1 was treated with DMAP, DCC, and (R)-/(S)-MPA reagent in CDCl3 at 0 °C. The (R)- and (S)-MPA esters of 1 (1-R and 1-S) were prepared, respectively, and the 1H NMR spectra of the resulting MPA esters were recorded in CDCl3. The differences in the chemical shifts (Δδ = δ
Penicacid F (2) was isolated as a white amorphous solid with the molecular formula C19H22O8 based on the HR-ESI-MS ion detected at m/z 401.1216 [M + Na]+. Analysis of the 1D NMR data (Table 1) of 2 revealed great similarity to those of 1. The major differences were the absence of an oxygenated methyl group and the appearance of an acetyl group (δH 2.02, δC 21.3, 7′-CH3; δC 170.3, C-7′) in 1. Slight changes were also observed for several proton and carbon signals (Table 1). The acetyl group was linked to C-2′ (δC 75.0) via an oxygen atom by the HMBC correlations from H-2′ to C-7′ (δC 170.3) and from 7′-CH3 to C-7′, and the planar structure of 2 was deduced according to the related key COSY and HMBC correlations shown in Fig. 2. Because 2 has only one chiral center at the same position as 1, according to the specific rotations ([α]25 D −17.3 for 1 and [α]25 D −16.2 for 2) and similar ECD data of 1 and 2 (see Fig. S33), the absolute configuration at C-2′ in 2 was assigned as S.
Penicacid G (3) was isolated as an amorphous solid with the molecular formula C18H22O7 established by HR-ESI-MS which displayed a peak at m/z 373.1255 [M + Na]+ (Calcd. for C18H22O7Na, 373.1258). The 1H and 13C NMR data of 3 closely resembled those of the known compound 4′-hydroxy-mycophenolic acid (5) [16] except for the appearance of a methoxyl group (δH 3.51, δC 51.1) in 3 instead of the hydroxyl group at C-6′ in 5. The location of the methoxy group at C-6′ in 3 was confirmed by the HMBC correlation from 6′-OCH3 to C-6′ (δC 171.3). Then, the whole planar structure of 3 could be determined on the basis of the key COSY and HMBC data as shown in Fig. 2.
The absolute configuration of C-4′ in 3 was determined to be S by the Mosher’s method as shown in Fig. 3 (see also Figs. S28 and S29), and thus the structure of compound 3 was established as 4′S-hydroxy-6′-methoxy mycophenolic acid.
All the compounds were evaluated for their cytotoxicity against HL-60 cell line by MTT method and HCT-116, BEL-7402, MGC-803, SH-SY5Y and HO-8910 cell lines by SRB method. Compounds 4 and 6 showed potent cytotoxicity against all of the tested six cell lines, with IC50 values ranging from 1.69 to 12.98 μmol·L–1, while compounds 2 and 3 showed moderated cytotoxicity, with IC50 values ranging from 12.61 to 26.38 μmol·L–1 (Table 2). In addition, compounds 1−3 were evaluated for their antimicrobial activity against Escherichia coli, Staphylococcus aureus, Proteus species, Candida albicans, and Bacillus subtilis. Only compound 3 showed a weak activity against Proteus species.
| Comp. | IC50 (μmol·L–1) | |||||
| HCT-116 | BEL-7402 | MGC803 | SH-SY5Y | HL-60 | HO-8910 | |
| 1 | > 30 | > 30 | > 30 | > 30 | > 30 | > 30 |
| 2 | 22.48 | 18.72 | 19.20 | 26.38 | 18.15 | > 30 |
| 3 | 20.80 | 12.61 | 19.75 | 16.74 | 17.80 | > 30 |
| 4 | 5.88 | 3.83 | 3.31 | 3.90 | 1.69 | 3.88 |
| 5 | > 30 | > 30 | > 30 | > 30 | > 30 | > 30 |
| 6 | 11.88 | 6.52 | 7.91 | 12.98 | 7.50 | 9.11 |
| Doxa | 0.21 | 0.37 | 0.17 | 0.15 | 0.02 | 0.37 |
| a Dox stands for doxorubicin hydrochloride, which was used as a reference drug. | ||||||
In conclusion, three new mycophenolic acid derivatives (1−3), along with three known compounds (4−6) were isolated from the deep-sea-derived fungus Penicillium parvum HDN17-478. The absolute configurations of 1 and 3 were determined by the Mosher’s method, and the absolute configuration of 2 was elucidated on the basis of the specific rotation and ECD data. It’s worth mentioning that penicacids E (1) and F (2) provide the first example of mycophenolic acid derivatives with a double bond at C-3′/C-4′ position. Moreover, it is the first time to report the cytotoxicity of 4 and 6 on the BEL-7402, MGC-803, and HO-8910 cell lines.
Optical rotations were measured with a JASCO P-1020 digital polarimeter (JASCO Corporation, Tokyo, Japan). UV spectra were recorded on a Beckman DU 640 spectrophotometer (Beckman Coulter Inc., Brea, CA, USA). IR spectra were recorded on a Bruker tensor-27 spectrophotometer using KBr discs (Bruker Corporation, Billerica, MA, USA). NMR spectra were recorded on an Agilent 500 and 600 MHz DD2 spectrometer using solvent as an internal standard and chemical shifts were recorded as δ-values (Agilent Technologies Inc., Santa Clara, CA, USA). HR-ESI-MS and ESI-MS data were obtained using a Thermo Scientific LTQ Orbitrap XL mass spectrometer. CD spectra were measured on a JASCO J-715 spectropolarimeter (JASCO Corporation, Tokyo, Japan). Column chromatography (CC) were performed on silica gel (200−300 mesh, Qingdao Marine Chemical Inc., Qingdao, China). Analytical HPLC was conducted on an ODS column (YMC-Pack ODS-A, 4.6 mm × 250 mm, S-5 μm, 1 mL·min–1). Semi-preparative HPLC collection was performed on an ODS column (YMC-Pack ODS-A, 10 mm × 250 mm, 5 μm, 3 mL·min–1). Medium-pressure preparation liquid chromatography (MPLC) was performed on a Bona-Agela CHEETAHTM HP100 (Beijing Agela Technologies Co., Ltd., Beijing, China).
The fungal strain Penicillium parvum HDN17-478 was isolated from marine sediment collected at 2476 m depth in South China Sea and identified by the sequence of the ITS region (GenBank accession number MN749812) with 99.44% similarity to Penicillium parvum. The strains were deposited at the Ministry of Education of China, School of Medicine and Pharmacy, Ocean University of China, Qingdao, China.
Erlenmeyer flasks each (1 L) containing 300 mL of fermentation medium were directly inoculated with spores (Penicillium parvum HDN17-478). The flasks were cultured under the static conditions at room temperature for 30 days. The fermentation medium contained starch (4.0%), sucrose (4.0%), maltose (3.0%), sodium glutamate (0.2%), peptone (0.2%), yeast extract (0.1%), soybean powder (0.05%), KH2PO4 (0.05%), and MgSO4·7H2O (0.03%) dissolved in naturally collected seawater (Huiquan Bay, Yellow Sea, Qiangdao, China).
The Penicillium parvum HDN17-478 fermentation broth (40 L) was filtered through cheese cloth to separate the supernatant from the mycelia. The supernatant was extracted with EtOAc. The mycelia was macerated and extracted with methanol. All extracts were evaporated under reduced pressure and combined to give a crude gum (15.0 g).
The extract was separated by VLC on silica gel using a stepped gradient elution with petroleum ether/CH2Cl2 (V/V, 100∶0 to 0∶100) and CH2Cl2/CH3OH (V/V, 100∶0, 98∶2, 96∶4, 94∶6, 80∶20, 1∶1, 0∶100) to obtain eight fractions (Fraction 1 to Fraction 8). Fraction 3 (98∶2 CH2Cl2/CH3OH) was further subjected to an ODS column eluting with CH3OH/H2O (V/V, 0%, 20%, 30%, 40%, 50%, 60%, 70%, 80%, 90%, 100%) to give 23 fractions (Fr.3-1−Fr.3-23). Fraction 3-13 was separated by semi-preparative HPLC eluted with MeOH/H2O/TFA (V/V/V 50∶50∶0.01, 3 mL·min–1) to obtain compound 5 (3.2 mg, tR = 24 min). Fraction 3-19 was separated by semi-preparative HPLC eluted with MeOH/H2O/TFA (V/V/V 60∶40∶0.01, 3 mL·min–1) to obtain compound 2 (10.4 mg, tR = 16 min) and compound 3 (8.3 mg, tR = 20 min). Fraction 3-20 was separated by semi-preparative HPLC eluted with MeCN/H2O/TFA (V/V/V 32∶68∶0.01, 3 mL·min–1) to obtain compound 4 (12.0 mg, tR = 19 min). Fraction 3-23 was separated by semi-preparative HPLC eluted with MeOH/H2O/TFA (V/V/V 65∶35∶0.01, 3 mL·min–1) to obtain compound 6 (19.3 mg, tR = 22 min). Fraction 4 (96 : 4 CH2Cl2/CH3OH) was further separated by MPLC eluting with CH3OH/H2O (V/V, 0−5 min, 10%−90%, 5−65 min, 10%−100%, 25 mL·min–1) to give 11 fractions (Fr.4-1−Fr.4-11). Fraction 4-8 was separated by semi-preparative HPLC eluted with MeOH/H2O/TFA (V/V/V 57∶43∶0.01, 3 mL·min–1) to obtain compound 1 (6.4 mg, tR = 17 min).
Penicacid E (1): white amorphous solid; [α]25 D −17.3 (c 0.08, MeOH); UV (MeOH) λmax (log ε): 220 (2.92), 248 (0.97), 300 (0.43) nm; ECD (MeOH) λ (nm) (Δ ε) 218 (+0.53); IR (KBr) νmax: 3566, 1734, 1684, 1457, cm−1; for 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 373.1253 [M + Na]+ (Calcd. for C18H22O7Na, 373.1258).
Penicacid F (2): white amorphous solid; [α]25 D −16.2 (c 0.1, MeOH); UV (MeOH) λmax (log ε): 220 (2.92), 248 (0.97), 300 (0.43) nm; ECD (MeOH) λ (nm) (Δ ε) 218 (+0.54); IR (KBr) νmax: 3362, 1734, 1623, 1457, cm−1; for 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 401.1216 [M + Na]+ (Calcd. for C19H22O8Na, 401.1207).
Penicacid G (3): amorphous solid; [α]25 D −4.2 (c 0.3, MeOH); UV (MeOH) λmax (log ε): 220 (2.92), 248 (0.97), 300 (0.43) nm; IR (KBr) νmax: 3430, 1735, 1621, 1456, cm−1; for 1H and 13C NMR data see Table 1; HR-ESI-MS m/z 373.1255 [M + Na]+ (Calcd. for C18H22O7Na, 373.1258).
The microorganism suspension (198 μL, 106 cfu·mL–1) in autoclaved growth medium [(peptone (20 g·L–1), beef extract (3 g·L–1), NaCl (5 g·L–1) for bacteria; peptone (20 g·L–1), yeast extract (10 g·L–1), glucose (20 g·L–1) for Candida albicans)] was added to each well of 96-well plates. Solutions (40 mmol·L–1) of the compounds and positive drugs were made up in DMSO and dispensed into 96-well plates using the 2× microdilution method, to give 16 concentrations in the range of 200–0.006 μmol·L–1. Incubated at 28 °C for 24 h, and the lowest concentration that gave complete growth inhibition was recorded as the minimum inhibitory concentration (MIC). Ciprofloxacin was used as a positive control.
These biological evaluations were carried out as previously reported in reference [19].
We thank the analytical group of the State Key Laboratory of Marine Drugs, Chinese Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, for all spectra tests.
1D and 2D NMR, HRESIMS spectra of compounds 1−3 are available as Supporting Information, which can be requested by sending E-mails to the corresponding author.
| [1] |
Xu XY, Zhang XY, Nong XH, et al. Brevianamides and mycophenolic acid derivatives from the deep-sea-derived fungus Penicillium brevicompactum DFFSCS025 [J]. Mar Drugs, 2017, 15(2): 43. doi: 10.3390/md15020043
|
| [2] |
Patil NS, Desai SB, Singh A, et al. A process for purification of mycophenolic acid: India, 2009040828 [P]. 2009-04-02.
|
| [3] |
Bentley R. Mycophenolic acid: A one hundred year odyssey from antibiotic to immunosuppressant [J]. Chem Rev, 2000, 100(10): 3801-3825. doi: 10.1021/cr990097b
|
| [4] |
Zhang Q, Yang BY, Li FL, et al. Mycophenolic acid derivatives with immunosuppressive activity from the coral-derived fungus Penicillium bialowiezense [J]. Mar Drugs, 2018, 16(7): 230. doi: 10.3390/md16070230
|
| [5] |
Takao N, Mikio S, Kunio A, et al. Some biological properties of mycophenolic acid [J]. J Antibiot, 1969, 22(4): 165-169. doi: 10.7164/antibiotics.22.165
|
| [6] |
Williams RH, Lively DH, Delong DC, et al. Mycophenolic acid: antiviral and antitumor properties [J]. Japanese J Antibiot, 1968, 21(7): 463-464. doi: 10.7164/antibiotics.21.463
|
| [7] |
Skropeta D. Deep-sea natural products [J]. Nat Prod Rep, 2008, 25(6): 1131-1166. doi: 10.1039/b808743a
|
| [8] |
Skropeta D, Wei LQ. Recent advances in deep-sea natural products [J]. Nat Prod Rep, 2014, 31(8): 999-1025. doi: 10.1039/C3NP70118B
|
| [9] |
Pilkington LI. A chemometric analysis of deep-sea natural products [J]. Molecules, 2019, 24(21): 3942. doi: 10.3390/molecules24213942
|
| [10] |
Zhang ZZ, He XQ, Che Q, et al. Sorbicillasins A-B and scirpyrone K from a deep-sea-derived fungus Phialocephala sp. FL30r [J]. Mar Drugs, 2018, 16(7): 245. doi: 10.3390/md16070245
|
| [11] |
Zhang ZZ, He XQ, Wu GW, et al. Aniline-tetramic acids from the deep-sea-derived fungus Cladosporium sphaerospermum L3P3 cultured with the HDAC inhibitor SAHA [J]. J Nat Prod, 2018, 81(7): 1651-1657. doi: 10.1021/acs.jnatprod.8b00289
|
| [12] |
Zhang ZZ, He XQ, Zhang GJ, et al. Inducing secondary metabolite production by combined culture of Talaromyces aculeatus and Penicillium variabile [J]. J Nat Prod, 2017, 80(12): 3167-3171. doi: 10.1021/acs.jnatprod.7b00417
|
| [13] |
Zhang ZZ, He XQ, Liu CC, et al. Clindanones A and B and cladosporols F and G, polyketides from the deep-sea derived fungus Cladosporium cladosporioides HDN14-342 [J]. RSC Adv, 2016, 6: 76498-76504. doi: 10.1039/C6RA14640F
|
| [14] |
Sun CX, Ge XP, Shah M, et al. New glutamine-containing azaphilone alkaloids from deep-sea-derived fungus Chaetomium globosum HDN151398 [J]. Mar Drugs, 2019, 17(5): 253. doi: 10.3390/md17050253
|
| [15] |
Lu XH, Zheng ZH, Zhang H, et al. Two new members of mycophenolic acid family from Penicillium brevicompactum Dierckx [J]. J Antibiot, 2009, 62(9): 527-529. doi: 10.1038/ja.2009.54
|
| [16] |
Habib E, León F, Bauer JD, et al. Mycophenolic derivatives from Eupenicillium parvum [J]. J Nat Prod, 2008, 71(11): 1915-1918. doi: 10.1021/np8003497
|
| [17] |
Rovirosa J, Marrero DA, Darias J, et al. Secondary metabolites from marine Penicillium brevicompactum [J]. J Chil Chem Soc, 2006, 51(1): 775-778.
|
| [18] |
Latypov SK, Seco JM, Quiñoá E, et al. MTPA vs MPA in the determination of the absolute configuration of chiral alcohols by 1H NMR [J]. J Org Chem, 1996, 61(24): 8569-8577. doi: 10.1021/jo960719i
|
| [19] |
Du L, Zhu TJ, Liu HB, et al. Cytotoxic polyketides from a marine-derived fungus Aspergillus glaucus [J]. J Nat Prod, 2008, 71(11): 1837-1842. doi: 10.1021/np800303t
|
| [1] | ZHAN Rui, WANG Zhi-Chong, YIN Ben-Lin, LIU Ying, CHEN Ye-Gao. Novel 9, 10-dihydrophenanthrene derivatives from Eria bambusifolia with cytotoxicity aganist human cancer cells in vitro[J]. Chinese Journal of Natural Medicines, 2016, 14(8): 621-625. |
| [2] | TANG Jing-Jing, GENG Xiao-Ting, WANG Ya-Jing, ZHENG Tian-Yu, LU Jin-Rong, HU Rong. Synthesis and cytotoxicity evaluation of 3-amino-2-hydroxypropoxyisoflavone derivatives[J]. Chinese Journal of Natural Medicines, 2016, 14(6): 462-472. |
| [3] | LI Yong-Wei, QI Jin, ZHANG Yuan-Yuan, HUANG Zhen, KOU Jun-Ping, ZHOU Shui-Ping, ZHANG Yu, YU Bo-Yang. Novel cytotoxic steroidal glycosides from the roots of Liriope muscari[J]. Chinese Journal of Natural Medicines, 2015, 13(6): 461-466. |
| [4] | WU Hai-Yan, WANG Wei-Guang, DU Xue, YANG Jin, PU Jian-Xin, SUN Han-Dong. Six new cytotoxic and anti-inflammatory 11, 20-epoxy-ent-kaurane diterpenoids from Isodon wikstroemioides[J]. Chinese Journal of Natural Medicines, 2015, 13(5): 383-389. |
| [5] | SHAN Yan, HONG Ting, WANG Yan-Fei, ZHANG Nen-Ling, YU Bo, LU Yu, QIU Sheng-Xiang. Synthesis and cytotoxicity of longistylin C derivatives[J]. Chinese Journal of Natural Medicines, 2015, 13(4): 311-315. |
| [6] | FAN Qiong-Ying, YIN Xia, LI Zheng-Hui, LI Yan, LIU Ji-Kai, FENG Tao, ZHAO Bao-Hua. Mycophenolic acid derivatives from cultures of the mushroom Laetiporus sulphureu[J]. Chinese Journal of Natural Medicines, 2014, 12(9): 685-688. |
| [7] | CAO Xing-Fen, WANG Jun-Song, WANG Peng-Ran, KONG Ling-Yi. Triterpenes from the stem bark of Mitragyna diversifolia and their cytotoxic activity[J]. Chinese Journal of Natural Medicines, 2014, 12(8): 628-631. |
| [8] | XU Ran, XIE Hai-Qin, DENG Lu-Lu, ZHANG Jian-Xin, YANG Fu-Mei, LIU Jian-Hua, HAO Xiao-Jiang, ZHANG Yuan-Hu. A new bufadienolide with cytotoxic activity from the Chinese traditional drug Ch'an Su[J]. Chinese Journal of Natural Medicines, 2014, 12(8): 623-627. |
| [9] | WANG Lin, LIU An, ZHANG Fei-Long, Yeung John H. K., LI Xu-Qin, CHO Chi-Hin. Evaluation and SAR analysis of the cytotoxicity of tanshinones in colon cancer cells[J]. Chinese Journal of Natural Medicines, 2014, 12(3): 167-171. |
| [10] | Consolacion Y. Ragasa, Kimberly B. Cornelio. Triterpenes from Euphorbia hirta and their cytotoxicity[J]. Chinese Journal of Natural Medicines, 2013, 11(5): 528-533. |
| 1. | Wang, C., Wang, Y., Xu, H. et al. Study on secondary metabolites of marine-derived Penicillium sp. HD-1-1 | [海洋真菌 Penicillium sp. HD-1-1 次级代谢产物研究]. Chinese Traditional and Herbal Drugs, 2024, 55(7): 2142-2151. DOI:10.7501/j.issn.0253-2670.2024.07.002 | |
| 2. | Mo, T., Qin, Y., Zhang, Y. et al. Three New Antibacterial Mycophenolic Acid Derivatives from the Marine-Derived Fungus Penicillium sp. HN-66. Chemistry and Biodiversity, 2024. DOI:10.1002/cbdv.202401657 | |
| 3. | Zhang, J.-X., Zhang, B.-D., Shi, Y. et al. Penindolacid A, a new indole alkaloid from the marine-derived fungus Penicillium sp.. Magnetic Resonance in Chemistry, 2023, 61(9-10): 554-559. DOI:10.1002/mrc.5389 | |
| 4. | Nesterenko, L.E., Popov, R.S., Zhuravleva, O.I. et al. A Study of the Metabolic Profiles of Penicillium dimorphosporum KMM 4689 Which Led to Its Re-Identification as Penicillium hispanicum. Fermentation, 2023, 9(4): 337. DOI:10.3390/fermentation9040337 | |
| 5. | Amin, T., Karim, A.B., Oyshe, I.I. et al. Unlocking Nature’s Treasure Trove: Exploring Microorganisms for Novel Bioactives. Journal of Angiotherapy, 2023, 7(1) DOI:10.25163/angiotherapy.719345 | |
| 6. | Zhang, J., Zhang, B., Cai, L. et al. New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata. Marine Drugs, 2022, 20(12): 778. DOI:10.3390/md20120778 | |
| 7. | Carroll, A.R., Copp, B.R., Davis, R.A. et al. Marine natural products. Natural Product Reports, 2022, 39(6): 1122-1171. DOI:10.1039/d1np00076d | |
| 8. | Yu, J., Wang, J.-P., Liu, S.-F. et al. 7-Methoxy-13-dehydroxypaxilline: New indole diterpenoid from an endophytic fungus Penicillium sp. Nb 19. Natural Product Research, 2022, 38(1): 103-111. DOI:10.1080/14786419.2022.2107639 | |
| 9. | Wang, H.-N., Sun, S.-S., Liu, M.-Z. et al. Natural bioactive compounds from marine fungi (2017–2020). Journal of Asian Natural Products Research, 2022, 24(3): 203-230. DOI:10.1080/10286020.2021.1947254 | |
| 10. | Pilevneli, A.D., Ebada, S.S., Kaşkatepe, B. et al. Penicacids H-J, three new mycophenolic acid derivatives from the marine-derived fungus: Rhizopus oryzae. RSC Advances, 2021, 11(55): 34938-34944. DOI:10.1039/d1ra07196c | |
| 11. | LI, X.-W.. Chemical ecology-driven discovery of bioactive marine natural products as potential drug leads. Chinese Journal of Natural Medicines, 2020, 18(11): 837-838. DOI:10.1016/S1875-5364(20)60024-3 |
| No. | 1a | 2a | 3b | |||||
| δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) | |||
| 1 | 172.7, C | 173.2, C | 170.1, C | |||||
| 3 | 70.2, CH2 | 5.21, s | 70.3, CH2 | 5.20, s | 68.6, CH2 | 5.24, s | ||
| 3a | 145.3, C | 144.6, C | 145.8, C | |||||
| 4 | 117.0, C | 117.0, C | 115.9, C | |||||
| 5 | 164.1, C | 163.9, C | 162.6, C | |||||
| 6 | 119.8, C | 121.4, C | 122.1, C | |||||
| 7 | 154.2, C | 153.8, C | 152.7, C | |||||
| 7a | 107.1, C | 106.7, C | 106.9, C | |||||
| 8 | 11.9, CH3 | 2.17, s | 11.8, CH3 | 2.15, s | 11.0, CH3 | 2.08, s | ||
| 9 | 61.2, CH3 | 3.82, s | 61.2, CH3 | 3.75, s | 60.6, CH3 | 3.69, s | ||
| 1′ | 30.3, CH2 | 2.92/2.99, m | 22.5, CH2 | 3.42, t (6.9) | 22.0, CH2 | 3.30, d (6.9) | ||
| 2′ | 77.1, CH | 4.34, d (7.7) | 75.0, CH | 5.53, t (7.7) | 122.8, CH | 5.36, d (6.6) | ||
| 3′ | 140.8, C | 132.6, C | 136.8, C | |||||
| 3′-CH3 | 12.6, CH3 | 1.77, s | 12.3, CH3 | 1.82, s | 11.8, CH3 | 1.70, s | ||
| 4′ | 117.2, CH | 5.64, t (6.8) | 127.1, CH | 5.60, t (6.7) | 72.5, CH | 4.20, m | ||
| 5′ | 33.4, CH2 | 3.09, d (4.9) | 38.3, CH2 | 2.63/2.75, m | 41.0, CH2 | 2.32/2.41, m | ||
| 6′ | 172.8, C | 175.0, C | 171.3, C | |||||
| 6′-OCH3 | 52.1, CH3 | 3.68, s | 51.1, CH3 | 3.51, s | ||||
| 7′ | 170.3, C | |||||||
| 7′-CH3 | 21.3, CH3 | 2.02, s | ||||||
| a Recorded in CDCl3; b Recorded in DMSO-d6 | ||||||||
| Comp. | IC50 (μmol·L–1) | |||||
| HCT-116 | BEL-7402 | MGC803 | SH-SY5Y | HL-60 | HO-8910 | |
| 1 | > 30 | > 30 | > 30 | > 30 | > 30 | > 30 |
| 2 | 22.48 | 18.72 | 19.20 | 26.38 | 18.15 | > 30 |
| 3 | 20.80 | 12.61 | 19.75 | 16.74 | 17.80 | > 30 |
| 4 | 5.88 | 3.83 | 3.31 | 3.90 | 1.69 | 3.88 |
| 5 | > 30 | > 30 | > 30 | > 30 | > 30 | > 30 |
| 6 | 11.88 | 6.52 | 7.91 | 12.98 | 7.50 | 9.11 |
| Doxa | 0.21 | 0.37 | 0.17 | 0.15 | 0.02 | 0.37 |
| a Dox stands for doxorubicin hydrochloride, which was used as a reference drug. | ||||||